#Written by Jessie Bolin (adapted from KScales stars code) December '23.
library(terra)
library(tidyverse)
library(ncdf4)
library(tmap)
library(sf)
### Read the metadata of the file
ncdf4::nc_open("/Users/jessicabolin/Dropbox/ocean_temp_mth_2020_12.nc") -> meta
meta #this shows the dimensions, variables and attributes of the file. Intro to netCDF
An overview
Most ocean products come in the format of netCDF. This is the most common data format and file type in the atmosphere and ocean sciences; essentially all output from weather, climate and ocean models is gridded data stored as a series of netCDF files. Satellite data is also often provided in NetCDF format.
The data in a netCDF file is stored in the form of arrays. netCDFs can contain 1D, 2D, 3D or 4D data. For example…
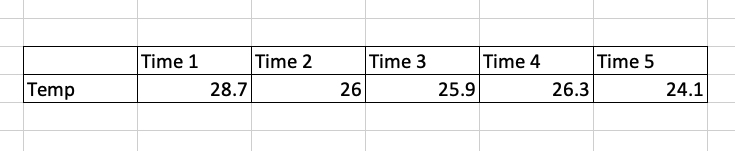
- Temperature varying over time at a location is stored as a one-dimensional array. You can think of it as a vector containing elements of the same data type (i.e. temperature values over time at a single longitude and latitude).
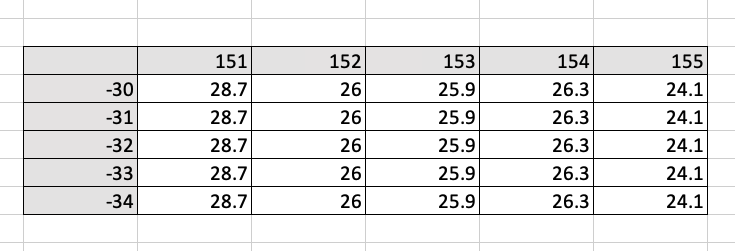
- An example of a 2-dimensional array is temperature over an area for a given time. Think of an excel spreadsheet, with a table of rows (longitude) and columns (latitude), and each element contains a temperature value for a given time.
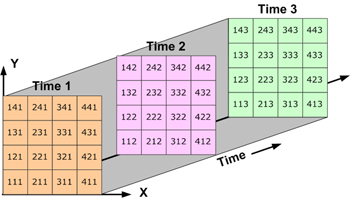
- Three-dimensional (3D) data is like temperature over an area varying with time. Think of multiple 2D arrays (lat/lon/temp) stacked on top of each other, with each ‘slice’ being a different time period (i.e., day).
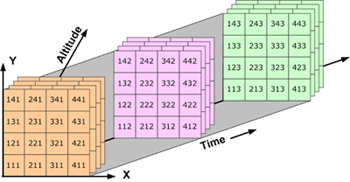
- Four-dimensional (4D) data is like temperature over an area varying with time and depth.
Working with .nc files
We use a combination of the ncdf4, terra, raster and stars packages to interact with .nc files in R. If dealing with climate model output, we also often use CDO via the system() function in R or through the terminal/shell.
We also use Panoply (free download) to quickly visualise data from netCDF files before interacting with them in R.
There are many ways to work with netCDFs in R, and a quick google will attest to that. Here are example workflows of using the terra and stars packages to read, manipulate and plot ocean temperature from a .nc file.
With terra
File /Users/jessicabolin/Dropbox/ocean_temp_mth_2020_12.nc (NC_FORMAT_NETCDF4):
5 variables (excluding dimension variables):
double average_DT[Time] (Chunking: [1]) (Compression: shuffle,level 1)
_FillValue: 1e+20
cell_methods: Time: mean
long_name: Length of average period
missing_value: 1e+20
units: days
double Time_bounds[nv,Time] (Chunking: [2,1]) (Compression: shuffle,level 1)
_FillValue: 1e+20
cell_methods: Time: mean
long_name: Time axis boundaries
missing_value: 1e+20
units: days
double average_T1[Time] (Chunking: [1]) (Compression: shuffle,level 1)
cell_methods: Time: mean
long_name: Start time for average period
units: days since 1979-01-01 00:00:00
missing_value: 1e+20
_FillValue: 1e+20
double average_T2[Time] (Chunking: [1]) (Compression: shuffle,level 1)
cell_methods: Time: mean
long_name: End time for average period
units: days since 1979-01-01 00:00:00
missing_value: 1e+20
_FillValue: 1e+20
float temp[xt_ocean,yt_ocean,st_ocean,Time] (Chunking: [300,300,1,1]) (Compression: shuffle,level 1)
long_name: Potential temperature
units: degrees C
valid_range: -32767
valid_range: 32767
missing_value: -32768
_FillValue: -32768
packing: 4
cell_methods: time: mean Time: mean
time_avg_info: average_T1,average_T2,average_DT
coordinates: geolon_t geolat_t
standard_name: sea_water_potential_temperature
6 dimensions:
Time Size:1 *** is unlimited ***
long_name: Time
units: days since 1979-01-01 00:00:00
cartesian_axis: T
calendar_type: GREGORIAN
calendar: GREGORIAN
bounds: Time_bounds
cell_methods: Time: mean
nv Size:2
xt_ocean Size:3600
long_name: tcell longitude
units: degrees_E
cartesian_axis: X
domain_decomposition: 1
domain_decomposition: 3600
domain_decomposition: 1
domain_decomposition: 1800
yt_ocean Size:1500
long_name: tcell latitude
units: degrees_N
cartesian_axis: Y
domain_decomposition: 1
domain_decomposition: 1500
domain_decomposition: 1
domain_decomposition: 150
st_ocean Size:51
long_name: tcell zstar depth
units: meters
cartesian_axis: Z
positive: down
edges: st_edges_ocean
st_edges_ocean Size:52
long_name: tcell zstar depth edges
units: meters
cartesian_axis: Z
positive: down
9 global attributes:
filename: TMP/ocean_temp_2020_12_01.nc.0000
NumFilesInSet: 20
grid_type: regular
grid_tile: N/A
history: Tue Sep 7 15:29:50 2021: ncap2 -O -s average_DT=average_T2-average_T1 -s Time_bounds(:,0)=average_T1 -s Time_bounds(:,1)=average_T2 ocean_temp_mth_2020_12.nc ocean_temp_mth_2020_12.nc
Mon Sep 6 14:26:43 2021: ncrcat -4 --dfl_lvl 1 --cnk_dmn Time,1 --cnk_dmn xt_ocean,300 --cnk_dmn yt_ocean,300 --cnk_dmn st_ocean,1 ocean_temp_2020_12_01.nc ocean_temp_2020_12_02.nc ocean_temp_2020_12_03.nc ocean_temp_2020_12_04.nc ocean_temp_2020_12_05.nc ocean_temp_2020_12_06.nc ocean_temp_2020_12_07.nc ocean_temp_2020_12_08.nc ocean_temp_2020_12_09.nc ocean_temp_2020_12_10.nc ocean_temp_2020_12_11.nc ocean_temp_2020_12_12.nc ocean_temp_2020_12_13.nc ocean_temp_2020_12_14.nc ocean_temp_2020_12_15.nc ocean_temp_2020_12_16.nc ocean_temp_2020_12_17.nc ocean_temp_2020_12_18.nc ocean_temp_2020_12_19.nc ocean_temp_2020_12_20.nc ocean_temp_2020_12_21.nc ocean_temp_2020_12_22.nc ocean_temp_2020_12_23.nc ocean_temp_2020_12_24.nc ocean_temp_2020_12_25.nc ocean_temp_2020_12_26.nc ocean_temp_2020_12_27.nc ocean_temp_2020_12_28.nc ocean_temp_2020_12_29.nc ocean_temp_2020_12_30.nc ocean_temp_2020_12_31.nc /g/data/gb6/BRAN/BRAN_tmp/daily/ocean_temp_2020_12.nc
NCO: netCDF Operators version 4.9.2 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
title: BRAN2020
catalogue_doi_url: http://dx.doi.org/10.25914/6009627c7af03
acknowledgement: BRAN is made freely available by CSIRO Bluelink and is supported by the Bluelink Partnership: a collaboration between the Australian Department of Defence, Bureau of Meteorology and CSIRO.# netCDF files don't often show 'normal' times/dates. Check the units and values of the time slice (note that we already know it should be December 2020)
meta$dim$Time$units #[1] "days since 1979-01-01 00:00:00"
meta$dim$Time$vals #[1] 15325.5
as.Date(meta$dim$Time$vals, origin = "1979-01-01") #[1] "2020-12-16".
# December 2020! Nice, we have confirmed the time.
# Read in the data
rast("/Users/jessicabolin/Dropbox/ocean_temp_mth_2020_12.nc") -> w
## print summary of w structure
w
# w has 51 layers, corresponding to depth slices (depth in metres)class : SpatRaster
dimensions : 1500, 3600, 51 (nrow, ncol, nlyr)
resolution : 0.1, 0.1 (x, y)
extent : -9.507305e-10, 360, -75, 75 (xmin, xmax, ymin, ymax)
coord. ref. :
source : ocean_temp_mth_2020_12.nc:temp
varname : temp (Potential temperature)
names : temp_~325.5, temp_~325.5, temp_~325.5, temp_~325.5, temp_~325.5, temp_~325.5, ...
unit : degrees C, degrees C, degrees C, degrees C, degrees C, degrees C, ... names(w) %>% head() #show the names of the first 6 layers
# we can see that each layer corresponds to a different depth slice.
# We already know that there is only one time field, hence why all times are the same[1] "temp_st_ocean=2.5_Time=15325.5" "temp_st_ocean=7.5_Time=15325.5"
[3] "temp_st_ocean=12.5_Time=15325.5" "temp_st_ocean=17.51539039611816_Time=15325.5"
[5] "temp_st_ocean=22.66702079772949_Time=15325.5" "temp_st_ocean=28.16938018798828_Time=15325.5"## This is a global field. We may want to crop it to our region of interest.
## filter to crop to region of interest by lon, lat (x, y)
w %>%
crop(c(151, 160, -33, -23)) -> x2
## slice to get a depth slice (here, top slice of 2.5m - the surface)
x3 <- x2[[1]]
# set crs
crs(x3) <- crs("epsg:4326") #this is lon/lat WGS84
x3class : SpatRaster
dimensions : 100, 90, 1 (nrow, ncol, nlyr)
resolution : 0.1, 0.1 (x, y)
extent : 151, 160, -33, -23 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source(s) : memory
name : temp_st_ocean=2.5_Time=15325.5
min value : 20.9396
max value : 27.8477 # Plot! the basic way
plot(x3, col = viridis::viridis(255))
maps::map("world", add = T, fill = T, col = "grey")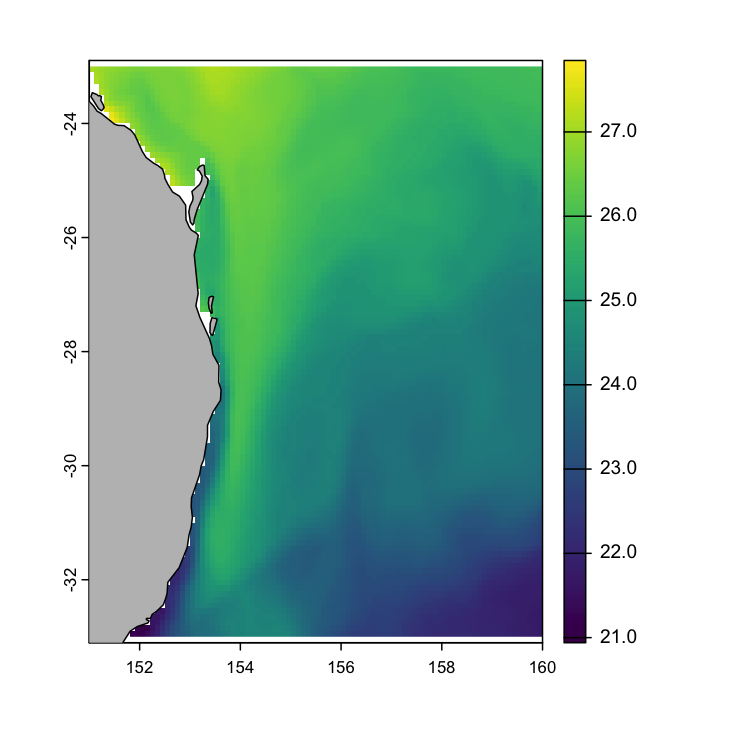
# the fancy way (with tmap)
aus <- rnaturalearth::ne_countries(country = 'Australia',
scale = 10,
returnclass = 'sf')
p1 <- tm_shape(x3) +
tm_raster(title = "SST",
style = "cont",
palette = viridis::viridis(255)) +
tm_graticules(ticks = T, lwd = 0.5,
n.x = 4, n.y = 4,
labels.size = 1,
col = "grey30") +
tm_legend(position = c("right", "top"),
legend.text.size = 1,
frame = T, frame.lwd = 0.001) +
tm_layout(main.title = "SST for December 2020 (BRAN)",
legend.title.size = 1.4,
bg.color = "white",
legend.frame = T,
fontfamily = "Arial Narrow") +
tm_shape(aus) +
tm_polygons()
p1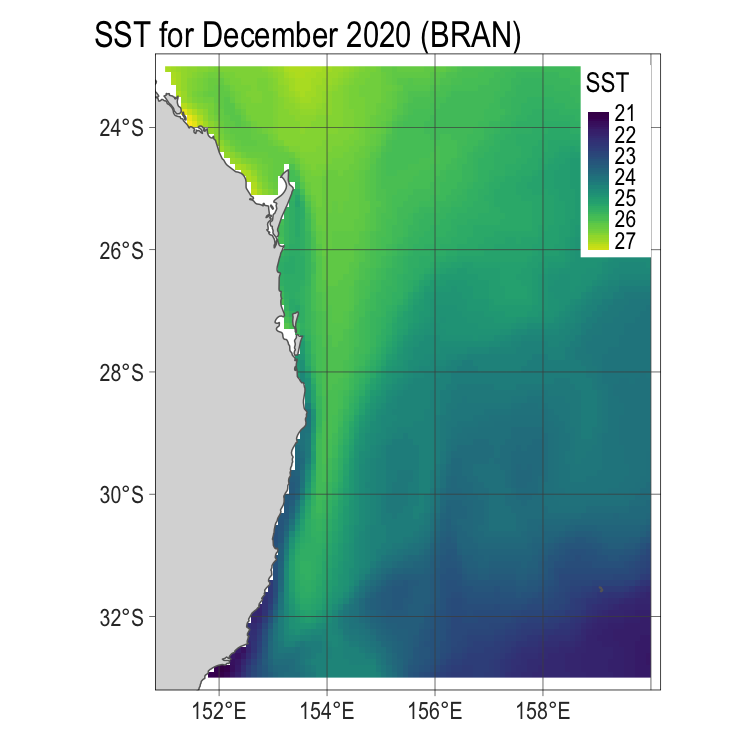
### EXTRACT point values from spatial coordinates (like fishing or animal sightings locations)
## random point (n=50) sample from within the bounding box of x3 (our study area)
pnt <- st_sample(st_as_sfc(st_bbox(x3)), 50) %>%
vect() #turn into spatvector so next function will work
pnt class : SpatVector
geometry : points
dimensions : 50, 0 (geometries, attributes)
extent : 151.2865, 159.5415, -32.92792, -23.364 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326) ## extract the values associated with these points
vals <- terra::extract(x3, pnt)
vals %>% head() ID temp_st_ocean=2.5_Time=15325.5
1 1 25.14601
2 2 25.51429
3 3 26.54028
4 4 25.02501
5 5 23.94579
6 6 NA## plot the physical data with point data overlaid. Add to p1, made earlier.
# need to convert the spatvector to a sf object in order to work with tmap.
p2 <- p1 +
tm_shape(pnt %>% st_as_sf()) +
tm_dots()
p2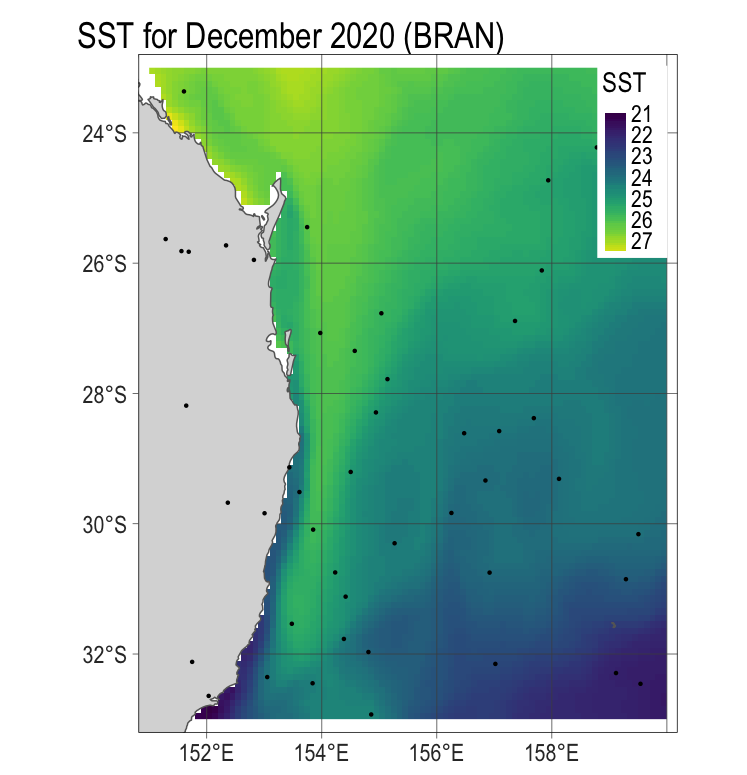
## we have some NAs as we have sampled over land. Let's remove those and replot.
vals2 <- na.omit(vals)
allvals <- pnt[vals2$ID] #only include the points that are not on land
allvals class : SpatVector
geometry : points
dimensions : 40, 0 (geometries, attributes)
extent : 151.6047, 159.5415, -32.92792, -23.364 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326) p3 <- p1 +
tm_shape(allvals %>% st_as_sf()) +
tm_dots()
p3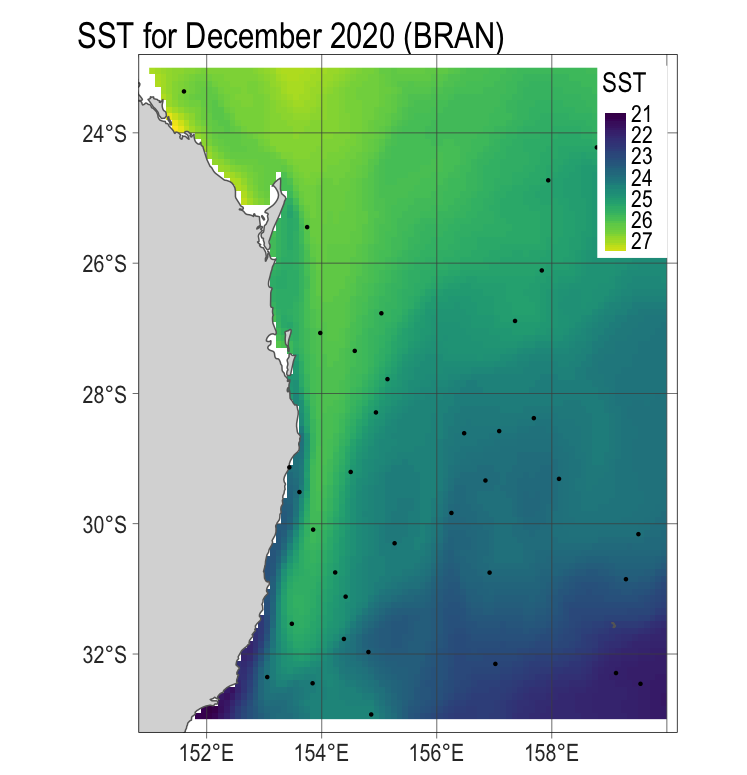
## we can convert our extracted temperature values to a data.frame, for visualisation, model fitting, etc.
temps <- as.data.frame(vals2)
# extract associated lat/lons
latlons <- pnt[vals2$ID] %>%
st_as_sf() %>%
st_coordinates() %>%
as.data.frame()
# combine
all_temps <- cbind(temps, latlons) %>% select(-ID)
names(all_temps) <- c("sst", "lon", "lat")
head(all_temps) sst lon lat
1 25.14601 157.8255 -26.11188
2 25.51429 158.8546 -24.15287
3 26.54028 151.6047 -23.36400
4 25.02501 154.9424 -28.29148
5 23.94579 156.8460 -29.33624
8 24.29825 155.2664 -30.29969With stars
Here is the above code rewritten in stars.
### Reading, plotting and manipulating physical data as NetCDF with package `stars`
### Written by Kylie L. Scales, March 2022
### Comprehensive help for `stars` and mapping to `raster` functions at https://r-spatial.github.io/stars/articles/stars1.html
### Physical data examples from ocean model BRAN2020 (BlueLink Reanalysis), CSIRO.
### Full info and data download at https://research.csiro.au/bluelink/outputs/data-access
library(stars)
library(cubelyr)
library(dplyr)
library(abind)
library(ggplot2)
library(viridis)
library(ggthemes)
## Download NetCDF from BRAN2020 from NCI data catalogue using THREDDS server
## https://dapds00.nci.org.au/thredds/catalog/gb6/BRAN/BRAN2020/catalog.html
## First, we'll use monthly ocean temperature data for December 2020, from:
## https://dapds00.nci.org.au/thredds/catalog/gb6/BRAN/BRAN2020/month/catalog.html
## scroll down to file `ocean_temp_mth_2020_12.nc` and click on filename
## click on HTTPServer link for a download of individual file through your browser
## save to your local drive (here, in ~/tmp/BRAN)
## We can automate download of multiple files from NCI, here using one as an example.
### READ the NetCDF file in using function read_ncdf(), and assign to an object, w
read_ncdf("/Users/jessicabolin/Dropbox/My Mac (Jessicas-MBP)/Desktop/Github/ocean_temp_mth_2020_12.nc") -> w
## print summary of w structure
w
## w has one attribute, temperature in °C
## w has four dimensions, xt_ocean (longitude), yt_ocean (latitude), st_ocean (depth), Time
## Time is a POSIXct object with meaningful dates as YYYY-MM-DD.
## There is only one time value as it's a single monthly field
## There are 51 depth slices, each with a corresponding value (depth in metres)
# select temp and drop singular time dimension ## NB. may need to keep if time matching point data later
x <- w %>% select(temp) %>% adrop()
x
## This is a global field. We may want to crop it to our region of interest.
## filter to crop to region of interest by lon, lat (x, y)
x
x %>%
filter(xt_ocean > 140, xt_ocean < 170, yt_ocean > -45, yt_ocean < -5) -> x2
x2
## slice to get a depth slice (here, top slice)
x2 %>%
slice(st_ocean,1) -> x3
x3
### PLOTTING using ggplot2()
## single depth layer (top slice)
p1 <- ggplot() +
geom_stars(data = x3, alpha = 0.8, downsample = c(1, 1, 1)) +
scale_fill_viridis() +
coord_equal() +
theme_map() +
theme(legend.position = "bottom") +
theme(legend.key.width = unit(2, "cm"))
p1
## several depth layers (here, top 12)
x2 %>%
slice(st_ocean,1:12) -> x4
x4
## open a new plotting window on a Mac. On a windows machine, use windows() instead
quartz()
ggplot() +
geom_stars(data = x4, alpha = 0.8, downsample = c(1, 1, 1)) +
facet_wrap("st_ocean") +
scale_fill_viridis() +
coord_equal() +
theme_map() +
theme(legend.position = "bottom") +
theme(legend.key.width = unit(2, "cm"))
### EXTRACT point values from spatial coordinates (like fishing or animal sightings locations)
## random point sample from within the bounding box of x3 (our study area)
pnt <- st_sample(st_as_sfc(st_bbox(x3)),50)
pnt
## extract the values associated with these point geometries
vals <- st_extract(x3, pnt)
vals
## plot the physical data with point data overlaid. Add to p1, made earlier.
p2 <- p1 +
geom_sf(data=pnt)
p2
## we have some NAs as we have sampled over land. Let's remove those and replot.
vals2 <- na.omit(vals)
p3 <- p1 +
geom_sf(data=vals2)
p3
## we can convert our extracted temperature values to a data.frame, for visualisation, model fitting, etc.
temps <- as.data.frame(vals2)
temps
## or, with longitude and latitude as separate columns, rather than point geometries
latlons <- st_coordinates(vals2) # Convert sf point geometries into a matrix of coordinates
latlons <- as.data.frame(latlons) # Convert from matrix to data frame
names(latlons) <- c("lon","lat")
temp2.5m <- as.vector(vals2$temp)
temps <- cbind(latlons, temp2.5m)
head(temps)